Featured Scientist: Robert D. Bradley PhD, Texas Tech University (TTU), Professor of Biological Sciences and Director of the Natural Science Research Laboratory, Museum of TTU.

Hometown: Grew up (first 18 years) in Diamond, MO. Lived various places as a student. Lived in Lubbock since 1994.
My Research: I study the relationships among mammals. I use genomic methods to answer conservation questions and taxonomic questions in mammals. I also work to improve genetic collections and natural history collections.
Research Goals: I would like to use genomics to develop “living” genetic databases that people can use for their research. These databases can be expanded every year. We can use them to provide real time information that can be used for conservation issues.
Career Goals: Develop the premier natural history collection at a university by enhancing the status of the TTU Natural Science Research Laboratory.
Hobbies: Ranching, hunting, and being a grandpa.
Favorite Thing About Science: Discovery of new information. Describing new species!
Scientist Upbringing: I knew nothing about being a scientist until I took a mammalogy class in 1982. I was “recruited” into a master’s research program and that eye-opening experience led to a career in science. I remember telling my professor (back in 1982) – “I can’t believe they pay you to do this… I would do it for free!”
My Team: I have published over 200 papers and all but two (book reviews) have included multiple authors, so collaborations are important to me. I led the field trip that collected the samples for the research described here. My friend (Hamilton) generated the chromosome data, and my PhD student (Mendez-Harclerode) did the DNA sequencing. We all worked together to write the paper.
Organism of Study: Baker’s small toothed harvest mouse (Reithrodontomys bakeri)

Field of Study: My primary field of study includes systematics, molecular evolution, and phylogenetics. This means that I study how the DNA of certain species changes over time and try to figure out how those changes lead to the development of new species. I focus mostly on mammals, and especially rodents in the southwestern United States, Mexico, and Central America. I use many types of data to understand events that play a role in natural history. More recently, we have started working in the field of mammalian genomics. In this field, we look at specific genes and how they have changed over time to try to understand how new species develop, and how certain species are related to each other.
Check Out My Original Paper: “A new species of Reithrodontomys from Guerrero, Mexico”

Citation: Bradley R. D., Mendez- Harclerode F., Hamilton M.J., & Ceballos G. (2004). A new species of Reithrodontomys from Guerrero, Mexico. Occasional Papers. 231, p. 1-12.
Article written by: Adriana Gonzalez (She/Her), B.S. in Biology, 2023, Payton Pargo (She/Her), B.S. in Biology, 2023, Priyanka Ghosh (She/Her), B.S. in Biology, 2023, Matthew C. Isaac (He/Him), B.S. in Natural Resources Management, Conservation Science, 2023, Troy Nations (He/Him), B.S., in Microbiology, 2023, Catherine Gooch (She/Her), B.S. in Kinesiology, 2025. Student authors were undergraduate students at TTU at the time the article was written.
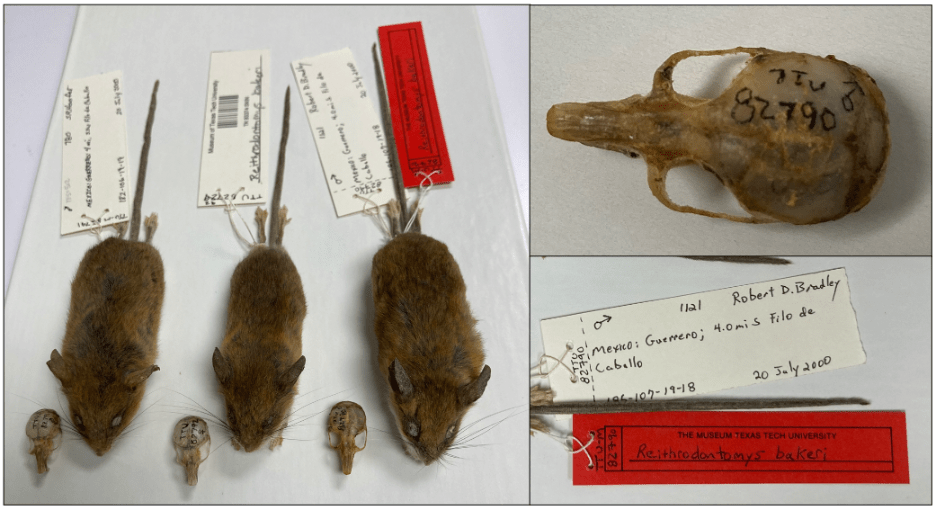
Research At A Glance: This research focused on small rodents that belong to the harvest mouse genus called Reithrodontomys. The authors trapped three harvest mice from two regions of Guerrero, Mexico. Their goal was to identify what specific Reithrodontomys species the captured mice belonged to. The authors used both genetic information and physical traits to classify the harvest mice. The genetic information that the authors collected were pictures of the mouse chromosomes and DNA sequences for each mouse. DNA sequences contain the instructions that the body uses to make proteins, which carry out most of the functions that our bodies need for everyday life. The instructions come in the form of a series of nucleotides. Nucleotides are the building blocks of DNA, and the specific order that they are in will determine the type of protein that the body will make. The authors compared the genetic information of the captured mice to other closely related species to see if they could find differences. These tests showed enough differences to suggest that the mice collected in Guerro, Mexico were a separate species. Another important set of information was the physical differences observed between these species. The authors measured the physical traits of the mice using natural history collections. Natural history collections contain preserved tissues, skins, and full body specimens that can be used for research. The authors used the collections to compare the mice from Guerrero, Mexico to other closely related mouse species. They measured physical traits, like ear length, tail length, and skull size. The authors found that the mice from Guerrero, Mexico had different sized skulls and ears when compared to the other species. Together, the genetic information and physical traits provided enough evidence for the authors to conclude that the Guerrero mice were a new species. The new species of harvest mouse was named Reithrodontomys bakeri, in honor of Robert S. Baker, a TTU Horn Professor.
Highlights: The purpose of this study was to correctly identify a species of harvest mouse collected in Guerrero, Mexico. Based on their appearance, the mice were initially thought to be a part of the Reithrodontomys microdon species. However, the mice from Guerrero, Mexico were located roughly 240 km south of the nearest recorded collection of other relatives (Figure 1). Therefore, the distance indicates that it is unlikely that they were R. microdon.

To properly classify the mice, researchers collected several types of data. First, the authors took pictures of the chromosomes of the three species collected in Guerrero, Mexico. They took five pictures for each mouse. The images showed that the mice from Guerrero, Mexico had identical chromosomes. This suggested that all three mice were the same species. Next, the authors collected DNA sequences. They isolated DNA from the mice collected from Guerrero, Mexico and mailed the DNA samples to a facility that could create DNA sequences. The authors also downloaded DNA sequences for 11 species of closely related mice from GenBank. GenBank is an online database where researchers can download and upload DNA sequences. The authors used the DNA sequences to create a phylogenetic tree. A phylogenetic tree shows the evolutionary relationships between species. The authors used several different statistical approaches to make the phylogenetic tree and each approach showed similar relationships between the species. The information from the phylogenetic trees suggested that the mice from Guerrero, Mexico were closely related to R. microdon (Figure 2). However, there were still enough genetic differences to suggest that they were a separate species.

The researchers also collected information on the physical traits of the mice. The authors measured the ears, tails, feet, and skulls of the same 11 species of closely related mice and compared them to the mice collected in Guerrero, Mexico. They then used a statistical test called an analysis of variance (ANOVA) to see if the physical traits of the mice from Guerrero, Mexico were different from the other mice. The authors found that the mice from Guerrero, Mexico had different ear and skull sizes than the other species. The location where the mice were captured, and the genetic and physical differences between the mice, led the authors to conclude that the mice from Guerrero, Mexico were a new species. This is how the species R. bakeri was discovered.
What My Science Looks Like: In this research, the authors used natural history collections to look at the physical traits of many mice. The information helped them determine that the mice they discovered in Mexico were their own species. Natural history collections are historical records of plants and animals (Figure 3). They often contain bones and tissues, but they can also include non-living samples, like rocks. These collections are important because they can help scientists understand what the world looked like in the past. Having a record from the past can also help scientists understand how species have changed over time. This information can be used for conservation and to understand the evolution of species over time.
The Big Picture: In this study, the authors identified and described a new species of harvest mouse. The research is important because the authors were able to identify a species that has been miscategorized in the past. The authors provide information on how the DNA and physical traits of these harvest mice differ from other species and this information can help scientists to correctly identify harvest mice in the future. The discovery of these mice also gives us insight into where the species originated and how they have evolved over time. One of the largest debates in taxonomy is what makes one species different from another? The authors used many techniques to distinguish between the species. These techniques can be adopted by others in the future.
Decoding the Language:
Analysis of variance (ANOVA): An ANOVA is a statistical tool designed to compare the average value of different groups to see if they are different from one another. In the context of this research, the author used an ANOVA to see if the average ear length for R. microdon was different from the average ear length of R. bakeri.
Chromosomes: Chromosomes are tiny structures that carry genetic information about an organism. They store this information in the form of genes, which are made of very long sequences of DNA. The research in this paper involved comparing the chromosomes of mouse species to look at the similarities between them.
Evolution: Evolution is a process in which heritable traits of a population change. It takes a long period of time for these changes to be noticeable, since they have to be passed down through many generations. But eventually, populations can look so different from their ancestors that they are considered a separate species from them. The discovery of the species R. bakeri is important because it provides insight to the evolution of harvest mice.
GenBank: Genbank is a publicly available website that shows nucleotide sequences and the proteins they code for. It is meant to be a source of comprehensive DNA sequence information for scientists to use. The author of this paper used GenBank to find the DNA sequences of 11 species of mice. This allowed them to see how genetically similar or different the mouse species were.
Genetic: Genetic means relating to heredity, which is the passing on of a trait or traits from parents to offspring. Different traits are caused by changes to sequences of DNA. In this research, the author looked at traits that were similar or different between the species of harvest mice and used them to determine how genetically similar the species were to each other.
Genus: A genus is a way to classify living organisms. It is usually the first word in the scientific name of a species. For example, Reithrodontomys bakeri is the scientific name for the newly discovered species of harvest mouse, where “bakeri” refers to the specific species and “Reithrodontomys” refers to the genus. The genus Reithrodontomys has many species of closely related mice.
Horn Professor: A Horn Professor is a title given to TTU faculty who have attained national and/or international recognition based on their scholarly achievements. Faculty are recognized as Horn Professors after an evaluation committee reviews letters of support from prominent scholars within their research field, reviews their record of scholarly publications, and evaluates their service to the university. It is considered the highest honor that a TTU Faculty member can receive.
Mammalian genomics: Mammalian genomics is the study of the genome in mammals. The genome is the entire set of genes in an organism, so research in mammalian genomics looks at all the different genes in mammal species. This allows scientists to determine evolutionary relationships between mammals.
Molecular evolution: Molecular evolution is the study of evolutionary changes in the DNA of a population. These changes happen over very long periods of time. The scientists in this study compared the DNA sequences of different harvest mice species. When they did this, they used a molecular evolution approach to understand the evolutionary relationships between the species.
Natural history collections: Natural history collections are specimens (or organisms) that a museum keeps. They allow scientists to use these specimens in their research. This allows scientists to make comparisons between species without having to collect the specimens in nature. The author of this research used natural history collections to gather physical information about various species of harvest mice in Mexico. This information was part of what allowed him to determine that R.bakeri was a different species from the others.
Nucleotides: Nucleotides are the building blocks of DNA. When the author of this paper compared the DNA sequences of mice species, the order of the nucleotides is what gave him the most information. This is because the specific order that nucleotides are in determines what information the DNA contains and what traits will be expressed.
Phylogenetic tree: A phylogenetic tree is a diagram that shows the evolutionary relationships between different species. The author of this study used the information he gathered to make a phylogenetic tree. This tree shows the evolutionary relationships between R. bakeri and other harvest mice species.
Sequencing: Sequencing is the process of determining the order of the nucleotides that make up DNA. Once this process has been done, the information is stored so that other scientists can use it in their research. The author of this paper used gene sequences to determine evolutionary relationships between R. bakeri and other harvest mice species.
Systematics/Taxonomy: Systematics is a field that deals with classification of species and taxonomy. Taxonomy is the science of classifying species into distinct groups based on how similar they are to each other. The author of this paper used information about the physical and genetic characteristics of harvest mice to classify R. bakeri as a separate species from the others.
Learn More:
A DNA sequencing fact sheet from the National Human Genome Research Institute that provides additional information on DNA sequencing.
An article from the Khan Academy that describes phylogenetic trees and explains how to read them.
A link to Dr. Robert Bradley’s lab website to learn more about his research at TTU.
An article to a more in-depth resource about ANOVA.
A link to the TTU Natural Science Laboratory, where the TTU specimen collections are located.
Synopsis edited by Dr. Rosario Marroquín-Flores, Texas Tech University, Department Biological Sciences.
Download this article here
Please take a survey to share your thoughts about the article!
